Authors: Eva Gato, Ignacio Pedro Constanso, Ana Candela, Fátima Galán, Bruno Kotska Rodiño-Janeiro, Manuel J. Arroyo, Gema Méndez, Luis Mancera, Tyler Alioto, Marta Gut, Ivo Gut, Miguel Álvarez-Tejado, Belén Rodríguez-Sánchez, Germán Bou, Marina Oviaño
DOI: 10.1128/JCM.00800-21
We present the latest published paper at JCM as a result of our collaboration with Microbiology Service from Hospital Universitario A Coruña. A project funding by SEIMC and carried out on CLOVER MS Data Analysis Software.
Abstract:
The increasing emergence of carbapenemase-producing Klebsiella pneumoniae (CPK) is a global health alarm. Rapid methods that require minimum sample preparation and rapid data analysis are urgently required. MALDI-TOF MS has recently been used by clinical laboratories for identification of antibiotic resistant bacteria; however, discrepancies have arisen regarding biological and technical issues. The aim of this study was to standardize an operating procedure and data analysis for identification of CPK by MALDI-TOF MS. To evaluate this approach, a series of 162 K. pneumoniae (112 CPK and 50 non CPK), were processed in the MALDI BioTyper system (Bruker Daltonik, Germany) following a standard operating procedure. The study was conducted in two stages, the first denominated the “Reproducibility stage” and the second, “CPK identification”. The first stage was designed to evaluate the biological and technical variation associated with the entire analysis of CPK and the second stage, to assess the final accuracy of MALDI-TOF for the identification of CPK. Therefore, we present an improved MALDI-TOF MS data analysis pipeline using neural network analysis implemented in Clover MS data analysis software (Clover Biosoft, Spain), that is designed to reduce variability, guarantee inter-laboratory reproducibility and maximize the information selected from the bacterial proteome. Using the Random Forest (RF) algorithm, 100% of CPK producing isolates were correctly identified when all the peaks in the spectra were selected as input features and TIC normalization was applied. Thus, we have demonstrated that real-time direct tracking of CPK is possible using MALDI-TOF MS.
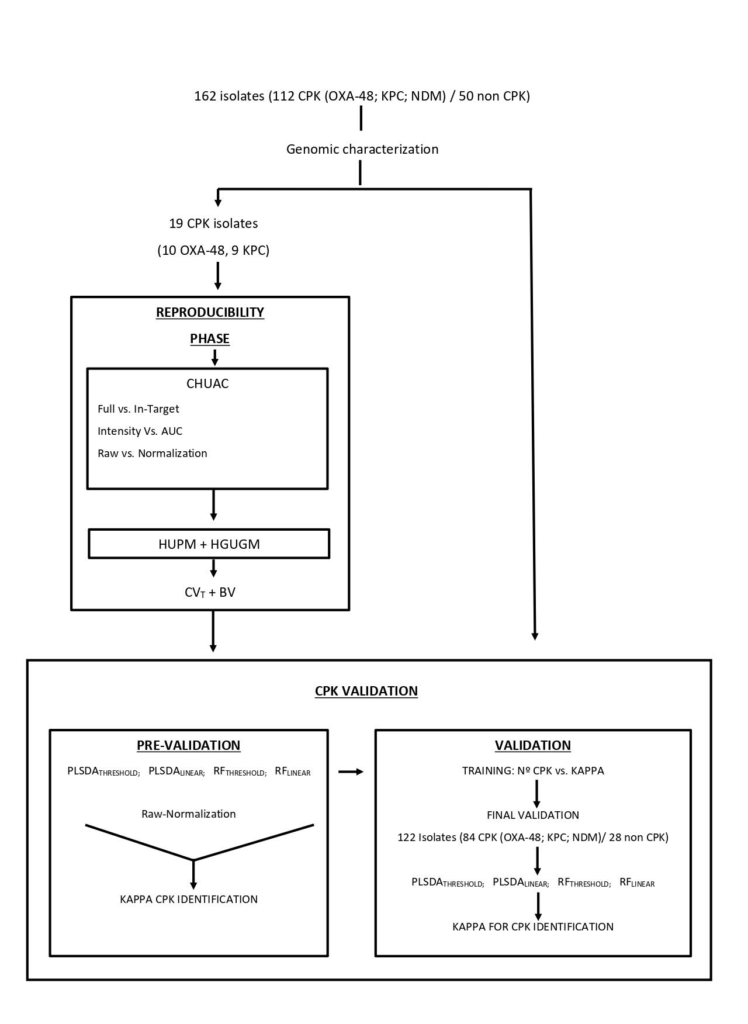
FULL ARTICLE:
