Author: Manuel J. Arroyo Pulgar
Using a machine-learning algorithm, MIT researchers have identified a powerful new antibiotic compound. In laboratory preliminary test, the drug killed many of the world’s most problematic disease-causing bacteria, including Acinetobacter baumannii and Enterobacteriaceae. To understand the relevance of such discovery, these bacteria are two of three high-priority pathogens that the WHO ranks as ‘critical’ for new antibiotics target.
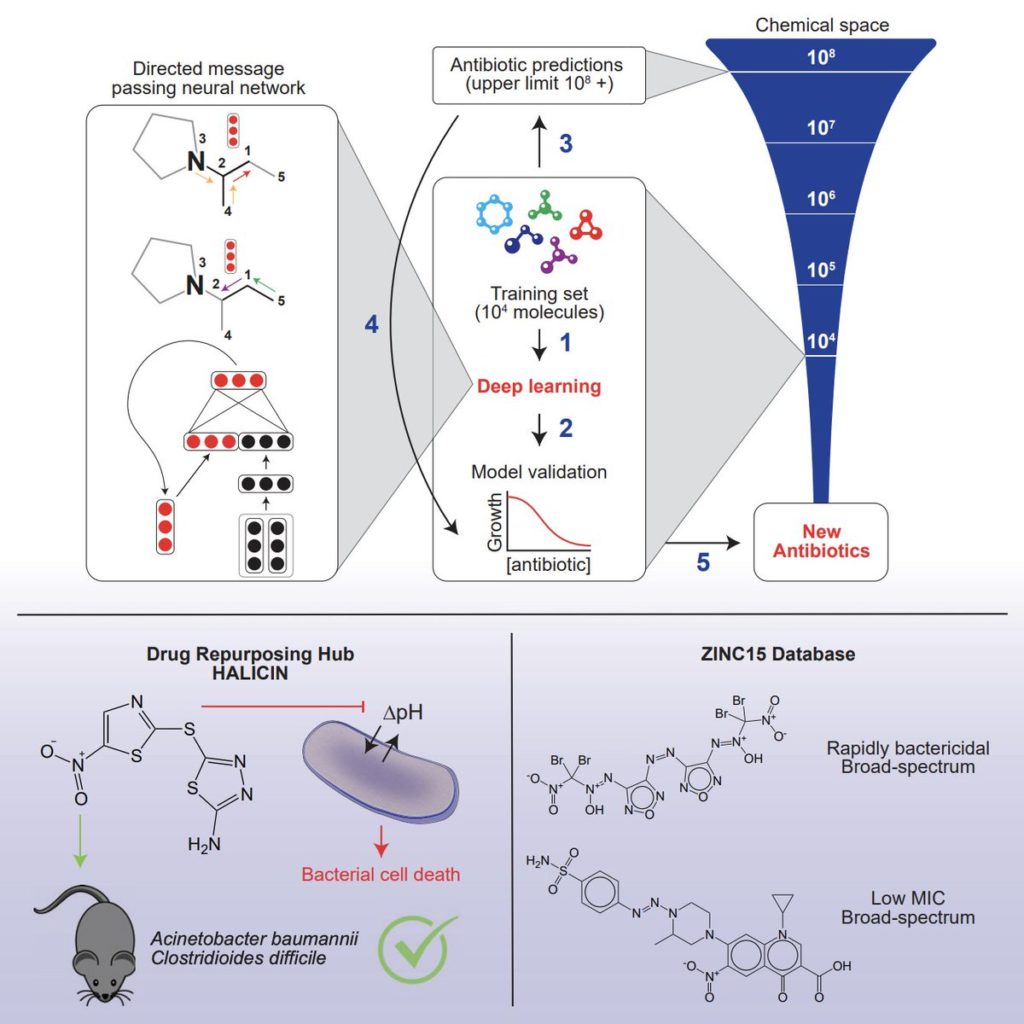
The computer model, which can screen more than 100 million chemical compounds in a matter of days, is designed to pick out potential antibiotics that kill bacteria using different mechanisms than those of existing drugs. To do this, researchers fed the program input with the atomic and molecular features of drugs and natural compounds, and how well or not the substance blocked the growth of the E. coli bacteria.
Once the model was trained, the researchers tested it on a library of about 6,000 compounds. The algorithm assessed the compounds and came up with some promising antibiotics. One of these molecules that was predicted to have strong antibacterial activity was called Halicin after the fictional artificial intelligence system from the movie 2001: A Space Odyssey.
This drug was previously investigated as possible diabetes drug, but it is the first time that an antibiotic was tested against microbial resistant bacteria. Researchers tested its activity and found that it was able to kill lots of infections that are resistant to conventional treatments, including Clostridium difficile, A. baumannii and Mycobacterium tuberculosis.
Preliminary studies suggest that Halicin kills bacteria by disrupting their ability to maintain an electrochemical gradient across their cell membranes. This gradient is necessary, among other functions, to produce ATP (molecules that cells use to store energy), so if the gradient breaks down, cells pass away. This type of killing mechanism could be difficult for bacteria to develop resistance to: When you’re dealing with a molecule that likely associates with membrane components, a cell can’t necessarily acquire a single mutation or a couple of mutations to change the chemistry of the outer membrane. Mutations like that tend to be far more complex to acquire evolutionarily, says Jonathan Strokes, a postdoc at MIT and the Broad Institute of MIT and Harvard and first author of the paper.
Other authors of the study now want to use the algorithm to find antibiotics that are more selective in the bacteria they kill. This would mean that taking the antibiotic would only kill the bugs that cause an infection, and not all the healthy bacteria that live in the gut. More ambitiously, the scientists aim to use the algorithm to design potent new antibiotics from scratch.
REFERENCE:
Stokes J. M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N. M., MacNair C. R., French S., Carfrae L. A., Bloom-Ackerman Z., Tran V. M., Chiappino-Pepe A., Badran A. H., Andrews I. W., Chory E. J., Church G. M., Brown E. D., Jaakkola T. S., Barzilay R. & Collins J. J. | 2020 | A Deep Learning Approach to Antibiotic Discovery. | Cell, 180(4), 688-702.e13. https://doi.org/10.1016/j.cell.2020.01.021
SOURCE NEWS:
http://news.mit.edu/2020/artificial-intelligence-identifies-new-antibiotic-0220
